DNN (Deep Neural Network)
neuralnet, nnet, deepnet
Neural Network
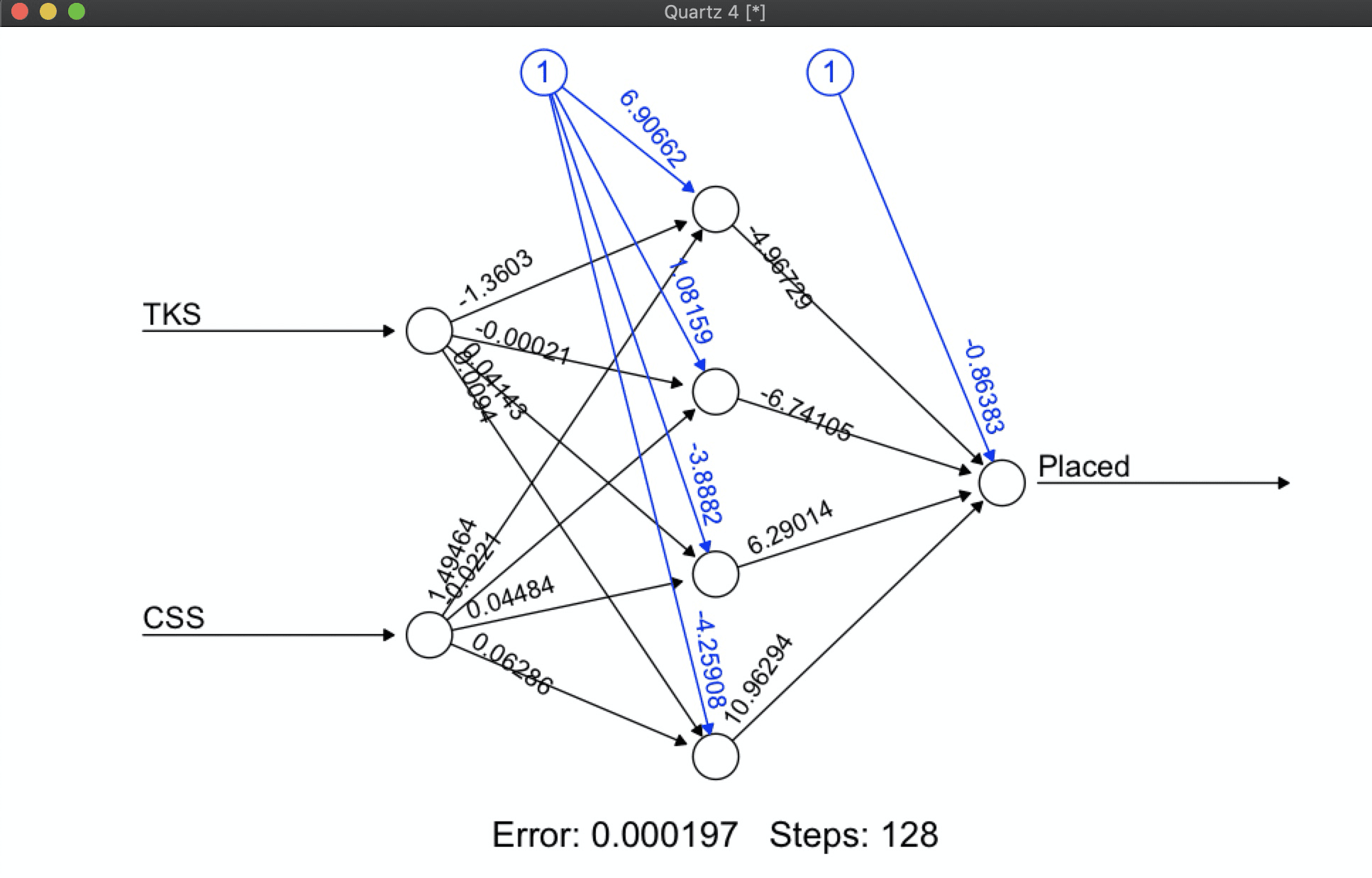
library(neuralnet) ### Loading Data --- --- --- --- --- --- --- --- --- --- --- --- --- --- ---- # creating train dataset ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ TKS <- c(20,10,30,20,80,30) #technical Knowledge Score -feature CSS <- c(90,20,40,50,50,80) #Communication Knowledge Score -feature Placed <- c( 1, 0, 0, 0, 1, 1) #Student Placed -binary label trnData <- data.frame(TKS,CSS,Placed) # creating test dataset ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ TKS <- c(30,40,85) CSS <- c(85,50,40) tstData <- data.frame(TKS,CSS)
# Network --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- ----
model <- neuralnet( Placed ~ TKS+CSS, data=trnData,
hidden=4, act.fct ="logistic", linear.output=F)
model %>% plot()
## Prediction using neural network -- --- --- --- --- --- --- --- --- --- ---- pred <- compute(model, tstData) pred.prob <- pred$net.result pred <- ifelse(pred.prob>0.5, 1, 0) pred
[,1] [1,] 1 [2,] 0 [3,] 1
classification
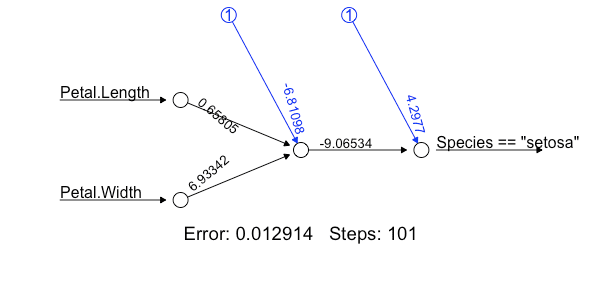
# Binary classification
nn <- neuralnet(formula= (Species=="setosa")~ Petal.Length + Petal.Width,
data=iris,
linear.output=FALSE)
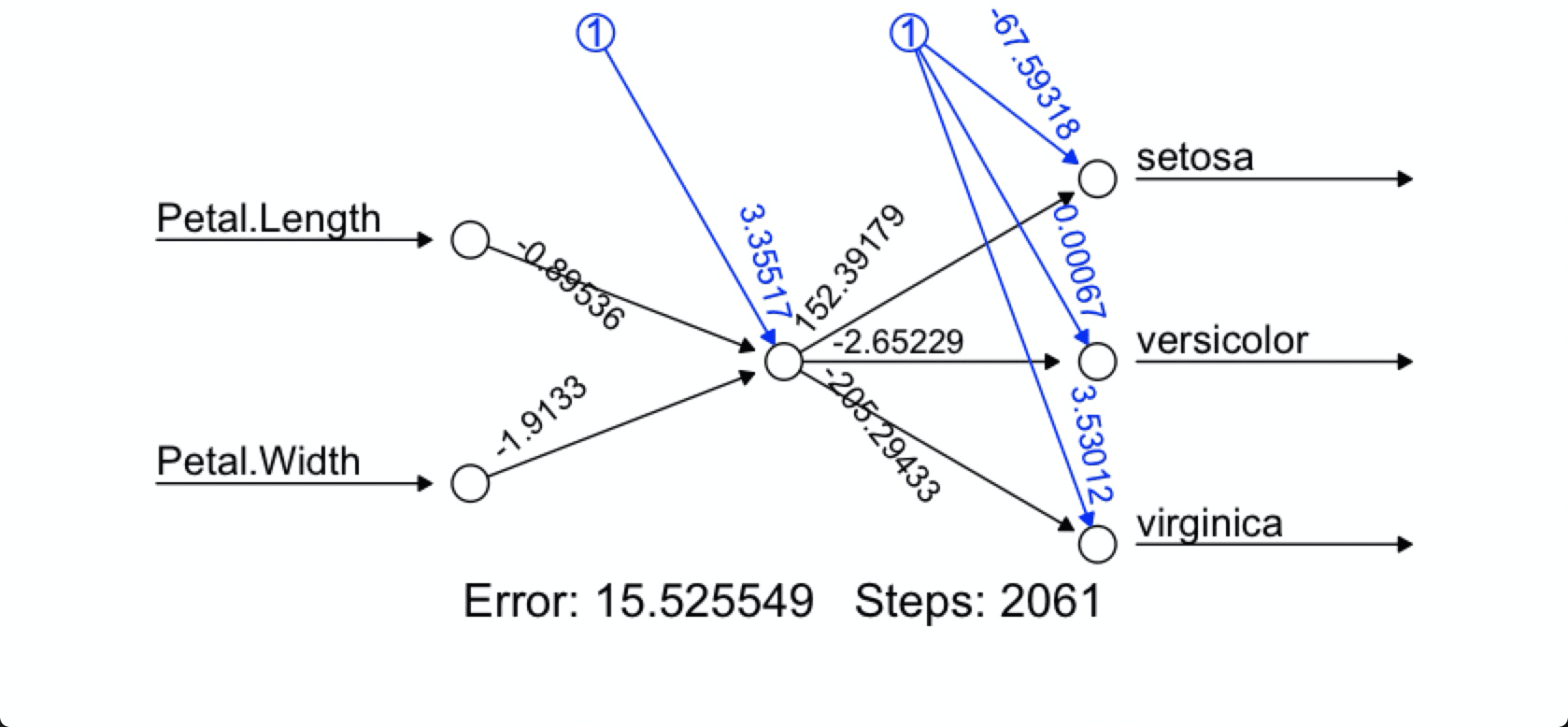
# Multiclass classification
nn <- neuralnet(formula= Species ~ Petal.Length + Petal.Width,
data=iris,
linear.output=FALSE)
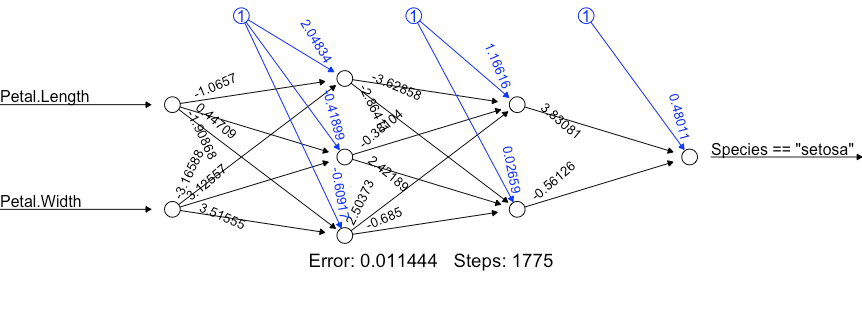
# Custom activation function
softplus <- function(x) log(1 + exp(x))
nn <- neuralnet(formula= (Species=="setosa")~ Petal.Length + Petal.Width,
data=iris,
linear.output=FALSE,
hidden = c(3, 2),
act.fct = softplus)
print(nn)
plot(nn)
library(neuralnet)
library(ggpmisc)
set.seed(2016)
# Load Data ---------------------------------------------------------------
attribute <- as.data.frame(sample(seq(-2,2, length=50), 50, replace=F), ncol=1)
response <- attribute^2
d0 <- cbind(attribute, response)
colnames(d0) <- c("attribute", "response")
d0 %>% head()
# ploting for TURE Value --------------------------------------------------
d0 %>% ggplot(aes(x=attribute, y=response)) + geom_point()
# Neuralnet regression ----------------------------------------------------
fit <- neuralnet(response~attribute, data=d0, hidden=c(3,3), threshold =0.01)
fit %>% summary
fit$net.result
# Predict ----------------------------------------------------------------
result <- cbind(d0, fit$net.result) %>% as.data.table()
colnames(result) <- c("attribute","response", "Pred")
result %>% ggplot(aes(x=attribute)) +
geom_point(aes(y=response)) + geom_point(aes(y=Pred), color="red")
result %>% ggplot(aes(x=response, y=Pred)) +
geom_point() + geom_smooth(method="lm", formula=y~x, se=F) +
stat_poly_eq(aes(label=..adj.rr.label..), formula=y~x, parse=T)
Resid <- result[, .(response-Pred)]
Resid %>% ggplot(aes(x=1:50, y=V1)) + geom_point()+
geom_smooth(method="lm", formula=y~x, se=F)
library('neuralnet')
library('Metrics') # for access model
library('ggpmisc')
### Load data ---------------------------------------------------------------
data("Boston", package="MASS")
dd <- Boston %>% data.table()
d1 <- scale(dd) %>% data.table() # x-mean/sd
keepcolumn <- c("crim","indus","nox","rm","age","dis","tax","ptratio","lstat","medv")
d2 <- d1[ , ..keepcolumn]
# Check NA
d2 %>% map_int( ~ sum(is.na(.)))
# Check Correlation
d2corr <- cor(d2)
d2melt <- melt(d2corr, varnames=c("x", "y"), value.name="Correlation")
d2melt <- d2melt[order(d2melt$Correlation), ]
d2melt %>% ggplot(aes(x=x, y=y)) + geom_tile(aes(fill=Correlation)) +
scale_fill_gradient2(
low="red", mid="white", high="steelblue",
guide=guide_colorbar(ticks=F, barheight=10),limits=c(-1, 1)) +
theme_minimal() + labs(x=NULL, y=NULL)
GGally::ggpairs(d2)
GGally::ggpairs(dd[ , ..keepcolumn])
### Model ---------------------------------------------------------------
set.seed(666)
train <- sample(x=1:dim(d2)[1], size=400, replace=F)
# fit <- neuralnet(formula= medv~crim+indus+nox+rm+age+dis+tax+ptratio+lstat,
# data= d2[train, ])
fit <- neuralnet(formula= medv~crim+indus+nox+rm+age+dis+tax+ptratio+lstat,
data= d2[train, ],
hidden=c(10,12,20), # No of Neurun for each Hidden layer
algorithm= "rprop+", # Resilient backpropagration with backtracking (instead of default 'backprop' with learningrate=0.01)
err.fct= "sse", # Error Function
act.fct= "logistic", # Activation Function
threshold= 0.1, # Activation Function threshold
linear.output= TRUE # Output layer neurun,
)
#pred <- compute(x=fit, covariate=d2[-train, 1:9])
pred <- predict(fit, newdata=d2[-train, 1:9])
### Assess Model ---------------------------------------------------------------
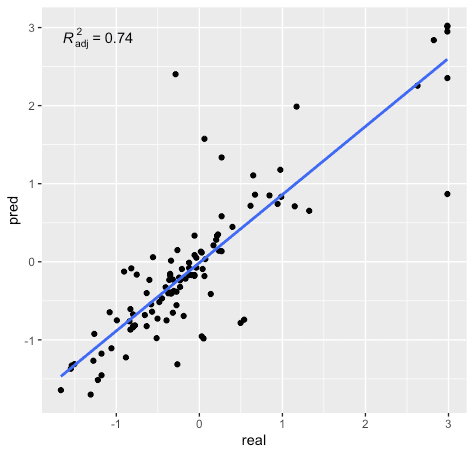
cor(d2[-train,][[10]] , pred)^2
#0.7458
mse(d2[-train,][[10]] , pred)
#0.2732
rmse(d2[-train,][[10]] , pred) # sqrt(mse)
#0.5227
RealvsPred <- cbind(d2[-train,][[10]] , pred) %>% data.table()
names(RealvsPred) <- c("real", "pred")
RealvsPred %>% ggplot(aes(x=real, y=pred)) + geom_point() +
geom_smooth(method="lm", formula=y~x, se=F) + stat_poly_eq(aes(label=..adj.rr.label..), formula=y~x, parse=T)
library(deepnet)
xx <- d2[train, 1:9] %>% as.matrix()
yy <- d2[train, 10] %>% as.matrix()
fit <- nn.train(x=xx, y=yy,
initW=NULL, initB=NULL, # initial weights, bias
hidden=c(10,12,20),
learningrate= 0.58, # for Backprop
momentum=0.74, # 경사하강 업데이틍에서 과거 결사들에 대한 가중치 평균
learningrate_scale=1, # 학습률
activationfun="sigm",
output="linear",
numepochs=970,
batchsize=60,
hidden_dropout=0,
visible_dropout=0
)
pred <- nn.predict(fit, x=d2[-train, 1:9])